- Record: found
- Abstract: found
- Article: not found
Generation of a mouse mutant by oligonucleotide-mediated gene modification in ES cells
Read this article at
Abstract
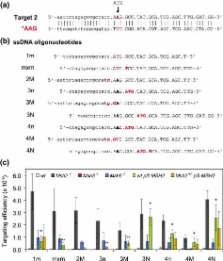
Oligonucleotide-mediated gene targeting is emerging as a powerful tool for the introduction of subtle gene modifications in mouse embryonic stem (ES) cells and the generation of mutant mice. However, its efficacy is strongly suppressed by DNA mismatch repair (MMR). Here we report a simple and rapid procedure for the generation of mouse mutants using transient down regulation of the central MMR protein MSH2 by RNA interference. We demonstrate that under this condition, unmodified single-stranded DNA oligonucleotides can be used to substitute single or several nucleotides. In particular, simultaneous substitution of four adjacent nucleotides was highly efficient, providing the opportunity to substitute virtually any given codon. We have used this method to create a codon substitution (N750F) in the Rb gene of mouse ES cells and show that the oligonucleotide-modified Rb allele can be transmitted through the germ line of mice.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Microsatellites in different eukaryotic genomes: survey and analysis.
- Record: found
- Abstract: found
- Article: not found
Altering the genome by homologous recombination.
- Record: found
- Abstract: found
- Article: not found
