- Record: found
- Abstract: found
- Article: found
Prioritized High-Confidence Risk Genes for Intellectual Disability Reveal Molecular Convergence During Brain Development
Read this article at
Abstract
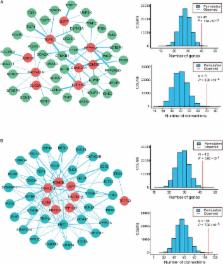
Dissecting the genetic susceptibility to intellectual disability (ID) based on de novo mutations (DNMs) will aid our understanding of the neurobiological and genetic basis of ID. In this study, we identify 63 high-confidence ID genes with q-values < 0.1 based on four background DNM rates and coding DNM data sets from multiple sequencing cohorts. Bioinformatic annotations revealed a higher burden of these 63 ID genes in FMRP targets and CHD8 targets, and these genes show evolutionary constraint against functional genetic variation. Moreover, these ID risk genes were preferentially expressed in the cortical regions from the early fetal to late mid-fetal stages. In particular, a genome-wide weighted co-expression network analysis suggested that ID genes tightly converge onto two biological modules (M1 and M2) during human brain development. Functional annotations showed specific enrichment of chromatin modification and transcriptional regulation for M1 and synaptic function for M2, implying the divergent etiology of the two modules. In addition, we curated 12 additional strong ID risk genes whose molecular interconnectivity with known ID genes ( q-values < 0.3) was greater than random. These findings further highlight the biological convergence of ID risk genes and help improve our understanding of the genetic architecture of ID.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability.
- Record: found
- Abstract: found
- Article: not found
Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders.
- Record: found
- Abstract: found
- Article: not found