- Record: found
- Abstract: found
- Article: found
A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life
Read this article at
Abstract
Background
COG0523 proteins are, like the nickel chaperones of the UreG family, part of the G3E family of GTPases linking them to metallocenter biosynthesis. Even though the first COG0523-encoding gene, cobW, was identified almost 20 years ago, little is known concerning the function of other members belonging to this ubiquitous family.
Results
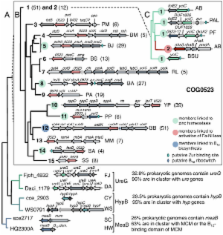
Based on a combination of comparative genomics, literature and phylogenetic analyses and experimental validations, the COG0523 family can be separated into at least fifteen subgroups. The CobW subgroup involved in cobalamin synthesis represents only one small sub-fraction of the family. Another, larger subgroup, is suggested to play a predominant role in the response to zinc limitation based on the presence of the corresponding COG0523-encoding genes downstream from putative Zur binding sites in many bacterial genomes. Zur binding sites in these genomes are also associated with candidate zinc-independent paralogs of zinc-dependent enzymes. Finally, the potential role of COG0523 in zinc homeostasis is not limited to Bacteria. We have predicted a link between COG0523 and regulation by zinc in Archaea and show that two COG0523 genes are induced upon zinc depletion in a eukaryotic reference organism, Chlamydomonas reinhardtii.
Conclusion
This work lays the foundation for the pursuit by experimental methods of the specific role of COG0523 members in metal trafficking. Based on phylogeny and comparative genomics, both the metal specificity and the protein target(s) might vary from one COG0523 subgroup to another. Additionally, Zur-dependent expression of COG0523 and putative paralogs of zinc-dependent proteins may represent a mechanism for hierarchal zinc distribution and zinc sparing in the face of inadequate zinc nutrition.
Related collections
Most cited references134
- Record: found
- Abstract: found
- Article: not found
WebLogo: a sequence logo generator.
- Record: found
- Abstract: found
- Article: not found
Jalview Version 2—a multiple sequence alignment editor and analysis workbench
- Record: found
- Abstract: not found
- Article: not found