- Record: found
- Abstract: found
- Article: found
Dew inspired breathing-based detection of genetic point mutation visualized by naked eye
research-article
Liping Xie
1
,
2 ,
Tongzhou Wang
1 ,
Tianqi Huang
1 ,
Wei Hou
4 ,
Guoliang Huang
b
,
1 ,
Yanan Du
a
,
1
,
3
09 September 2014
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
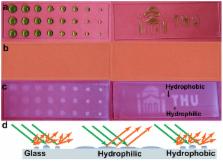
A novel label-free method based on breathing-induced vapor condensation was developed for detection of genetic point mutation. The dew-inspired detection was realized by integration of target-induced DNA ligation with rolling circle amplification (RCA). The vapor condensation induced by breathing transduced the RCA-amplified variances in DNA contents into visible contrast. The image could be recorded by a cell phone for further or even remote analysis. This green assay offers a naked-eye-reading method potentially applied for point-of-care liver cancer diagnosis in resource-limited regions.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Mutation detection and single-molecule counting using isothermal rolling-circle amplification.
P M Lizardi, X. Huang, Z. Zhu … (1998)
- Record: found
- Abstract: found
- Article: not found