- Record: found
- Abstract: found
- Article: found
The nucleoid occlusion factor Noc controls DNA replication initiation in Staphylococcus aureus
Read this article at
Abstract
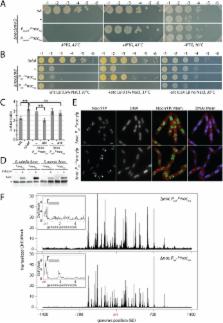
Successive division events in the spherically shaped bacterium Staphylococcus aureus are oriented in three alternating perpendicular planes. The mechanisms that underlie this relatively unique pattern of division and coordinate it with chromosome segregation remain largely unknown. Thus far, the only known spatial regulator of division in this organism is the nucleoid occlusion protein Noc that inhibits assembly of the cytokinetic ring over the chromosome. However, Noc is not essential in S. aureus, indicating that additional regulators are likely to exist. To search for these factors, we screened for mutants that are synthetic lethal with Noc inactivation. Our characterization of these mutants led to the discovery that S. aureus Noc also controls the initiation of DNA replication. We show that cells lacking Noc over-initiate and mutations in the initiator gene dnaA suppress this defect. Importantly, these dnaA mutations also partially suppress the division problems associated with Δ noc. Reciprocally, we show that over-expression of DnaA enhances the over-initiation and cell division phenotypes of the Δ noc mutant. Thus, a single factor both blocks cell division over chromosomes and helps to ensure that new rounds of DNA replication are not initiated prematurely. This degree of economy in coordinating key cell biological processes has not been observed in rod-shaped bacteria and may reflect the challenges posed by the reduced cell volume and complicated division pattern of this spherical pathogen.
Author summary
The mechanisms by which bacteria coordinate cell division with chromosome replication and segregation remain poorly understood. This coordination is particularly challenging in the spherical bacterium Staphylococcus aureus, which unlike rod-shaped bacteria, divides in three consecutive perpendicular division planes. The only known spatial regulator of division in S. aurues is the nucleoid occlusion protein Noc. In Bacillus subtilis, Noc has been shown to bind specific DNA sequences on the chromosome and block the assembly of the cell division apparatus over these sites. Because these binding sites are enriched in the origin-proximal portion of the chromosome and absent in the terminus region, Noc is thought to help coordinate cell division with chromosome segregation. Here, we report that S. aureus Noc protein not only plays a similar role in nucleoid occlusion, but also controls the initiation of DNA replication, thus providing an even tighter connection between cell division and chromosome biology than previously appreciated. This degree of economy in coordinating key cell biological processes may reflect the challenges posed by the small cell size and complicated division pattern of this spherical pathogen.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
NIH Image to ImageJ: 25 years of image analysis.

- Record: found
- Abstract: found
- Article: found
A Genetic Resource for Rapid and Comprehensive Phenotype Screening of Nonessential Staphylococcus aureus Genes

- Record: found
- Abstract: found
- Article: found