- Record: found
- Abstract: found
- Article: found
Inactivation Kinetics and Mechanical Gating of Piezo1 Ion Channels Depend on Subdomains within the Cap

Read this article at
SUMMARY
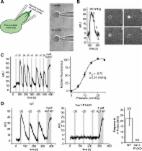
Piezo1 ion channels are activated by mechanical stimuli and mediate the sensing of blood flow. Although cryo-electron microscopy (cryo-EM) structures have revealed the overall architecture of Piezo1, the precise domains involved in activation and subsequent inactivation have remained elusive. Here, we perform a targeted chimeric screen between Piezo1 and the closely related isoform Piezo2 and use electrophysiology to characterize their inactivation kinetics during mechanical stimulation. We identify three small subdomains within the extracellular cap that individually can confer the distinct kinetics of inactivation of Piezo2 onto Piezo1. We further show by cysteine crosslinking that conformational flexibility of these subdomains is required for mechanical activation to occur and that electrostatic interactions functionally couple the cap to the extensive blades, which have been proposed to function as sensors of membrane curvature and tension. This study provides a demonstration of internal gating motions involved in mechanotransduction by Piezo1.
Graphical Abstract

In Brief
Lewis and Grandl combine a chimeric screen and cysteine crosslinking to identify small subdomains of the cap of mechanically activated Piezo1 ion channels that must have conformational flexibility for mechanical gating. They further show that electrostatic interactions couple one of these domains to the channel blade.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Piezos are pore-forming subunits of mechanically activated channels
- Record: found
- Abstract: found
- Article: not found
Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells.
- Record: found
- Abstract: found
- Article: found