- Record: found
- Abstract: found
- Article: found
High-density linkage mapping and evolution of paralogs and orthologs in Salix and Populus
Read this article at
Abstract
Background
Salix (willow) and Populus (poplar) are members of the Salicaceae family and they share many ecological as well as genetic and genomic characteristics. The interest of using willow for biomass production is growing, which has resulted in increased pressure on breeding of high yielding and resistant clones adapted to different environments. The main purpose of this work was to develop dense genetic linkage maps for mapping of traits related to yield and resistance in willow. We used the Populus trichocarpa genome to extract evenly spaced markers and mapped the orthologous loci in the willow genome. The marker positions in the two genomes were used to study genome evolution since the divergence of the two lineages some 45 mya.
Results
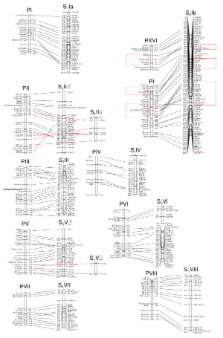
We constructed two linkage maps covering the 19 linkage groups in willow. The most detailed consensus map, S 1, contains 495 markers with a total genetic distance of 2477 cM and an average distance of 5.0 cM between the markers. The S 3 consensus map contains 221 markers and has a total genetic distance of 1793 cM and an average distance of 8.1 cM between the markers. We found high degree of synteny and gene order conservation between willow and poplar. There is however evidence for two major interchromosomal rearrangements involving poplar LG I and XVI and willow LG Ib, suggesting a fission or a fusion in one of the lineages, as well as five intrachromosomal inversions. The number of silent substitutions were three times lower (median: 0.12) between orthologs than between paralogs (median: 0.37 - 0.41).
Conclusions
The relatively slow rates of genomic change between willow and poplar mean that the genomic resources in poplar will be most useful in genomic research in willow, such as identifying genes underlying QTLs of important traits. Our data suggest that the whole-genome duplication occurred long before the divergence of the two genera, events which have until now been regarded as contemporary. Estimated silent substitution rates were 1.28 × 10 -9 and 1.68 × 10 -9 per site and year, which are close to rates found in other perennials but much lower than rates in annuals.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Populus: a model system for plant biology.
- Record: found
- Abstract: found
- Article: not found
Bioenergy from plants and the sustainable yield challenge.
- Record: found
- Abstract: found
- Article: not found