- Record: found
- Abstract: found
- Article: found
Loss of RNF43 Function Contributes to Gastric Carcinogenesis by Impairing DNA Damage Response
Read this article at
Abstract
Background & Aims
RING finger protein 43 (RNF43) is a tumor suppressor that frequently is mutated in gastric tumors. The link between RNF43 and modulation of Wingless-related integration site (WNT) signaling has not been shown clearly in the stomach. Because mutations in RNF43 are highly enriched in microsatellite-unstable gastric tumors, which show defects in DNA damage response (DDR), we investigated whether RNF43 is involved in DDR in the stomach.
Methods
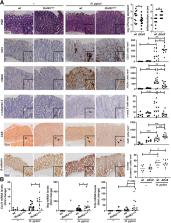
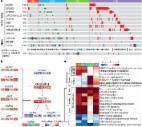
DDR activation and cell viability upon γ-radiation was analyzed in gastric cells where expression of RNF43 was depleted. Response to chemotherapeutic agents 5-fluorouracil and cisplatin was analyzed in gastric cancer cell lines and xenograft tumors. In addition, involvement of RNF43 in DDR activation was analyzed upon Helicobacter pylori infection in wild-type and Rnf43 ΔEx8 mice. Furthermore, a cohort of human gastric biopsy specimens was analyzed for RNF43 expression and mutation status as well as for activation of DDR.
Results
RNF43 depletion conferred resistance to γ-radiation and chemotherapy by dampening the activation of DDR, thereby preventing apoptosis in gastric cells. Upon Helicobacter pylori infection, RNF43 loss of function reduced activation of DDR and apoptosis. Furthermore, RNF43 expression correlated with DDR activation in human gastric biopsy specimens, and RNF43 mutations found in gastric tumors conferred resistance to DNA damage. When exploring the molecular mechanisms behind these findings, a direct interaction between RNF43 and phosphorylated H2A histone family member X (γH2AX) was observed.
Graphical abstract
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: found
Comprehensive molecular characterization of gastric adenocarcinoma
- Record: found
- Abstract: found
- Article: not found
The DNA-damage response in human biology and disease.
- Record: found
- Abstract: not found
- Article: not found
