- Record: found
- Abstract: found
- Article: found
Personalization of Logical Models With Multi-Omics Data Allows Clinical Stratification of Patients
Read this article at
Abstract
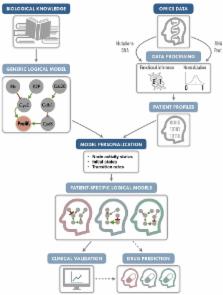
Logical models of cancer pathways are typically built by mining the literature for relevant experimental observations. They are usually generic as they apply for large cohorts of individuals. As a consequence, they generally do not capture the heterogeneity of patient tumors and their therapeutic responses. We present here a novel framework, referred to as PROFILE, to tailor logical models to a particular biological sample such as a patient tumor. This methodology permits to compare the model simulations to individual clinical data, i.e., survival time. Our approach focuses on integrating mutation data, copy number alterations (CNA), and expression data (transcriptomics or proteomics) to logical models. These data need first to be either binarized or set between 0 and 1, and can then be incorporated in the logical model by modifying the activity of the node, the initial conditions or the state transition rates. The use of MaBoSS, a tool based on Monte-Carlo kinetic algorithm to perform stochastic simulations on logical models results in model state probabilities, and allows for a semi-quantitative study of the model phenotypes and perturbations. As a proof of concept, we use a published generic model of cancer signaling pathways and molecular data from METABRIC breast cancer patients. For this example, we test several combinations of data incorporation and discuss that, with these data, the most comprehensive patient-specific cancer models are obtained by modifying the nodes' activity of the model with mutations, in combination or not with CNA data, and altering the transition rates with RNA expression. We conclude that these model simulations show good correlation with clinical data such as patients' Nottingham prognostic index (NPI) subgrouping and survival time. We observe that two highly relevant cancer phenotypes derived from personalized models, Proliferation and Apoptosis, are biologically consistent prognostic factors: patients with both high proliferation and low apoptosis have the worst survival rate, and conversely. Our approach aims to combine the mechanistic insights of logical modeling with multi-omics data integration to provide patient-relevant models. This work leads to the use of logical modeling for precision medicine and will eventually facilitate the choice of patient-specific drug treatments by physicians.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.
- Record: found
- Abstract: found
- Article: not found
Dynamical analysis of a generic Boolean model for the control of the mammalian cell cycle.
- Record: found
- Abstract: found
- Article: not found