- Record: found
- Abstract: found
- Article: found
CircadiOmics: circadian omic web portal
Read this article at
Abstract
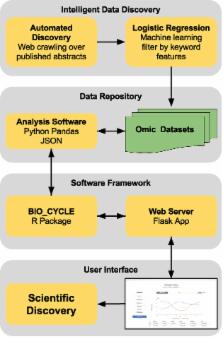
Circadian rhythms play a fundamental role at all levels of biological organization. Understanding the mechanisms and implications of circadian oscillations continues to be the focus of intense research. However, there has been no comprehensive and integrated way for accessing and mining all circadian omic datasets. The latest release of CircadiOmics ( http://circadiomics.ics.uci.edu) fills this gap for providing the most comprehensive web server for studying circadian data. The newly updated version contains high-throughput 227 omic datasets corresponding to over 74 million measurements sampled over 24 h cycles. Users can visualize and compare oscillatory trajectories across species, tissues and conditions. Periodicity statistics (e.g. period, amplitude, phase, P-value, q-value etc.) obtained from BIO_CYCLE and other methods are provided for all samples in the repository and can easily be downloaded in the form of publication-ready figures and tables. New features and substantial improvements in performance and data volume make CircadiOmics a powerful web portal for integrated analysis of circadian omic data.
Related collections
Most cited references22
- Record: found
- Abstract: found
- Article: not found
Transcriptional architecture and chromatin landscape of the core circadian clock in mammals.
- Record: found
- Abstract: found
- Article: not found
Molecular architecture of the mammalian circadian clock.
- Record: found
- Abstract: found
- Article: not found