- Record: found
- Abstract: found
- Article: found
ZEBRA: a hierarchically integrated gene expression atlas of the murine and human brain at single-cell resolution

Read this article at
Abstract
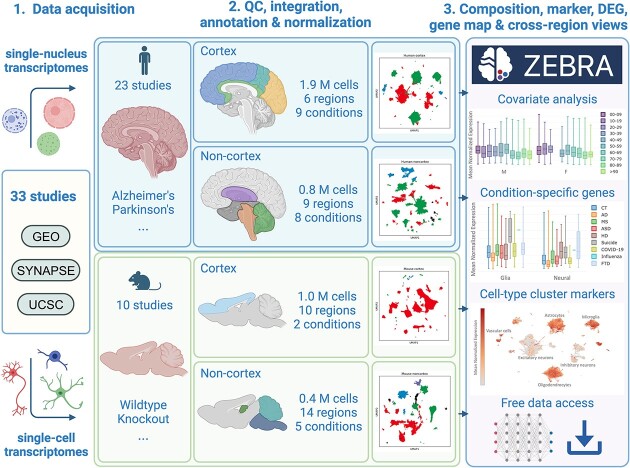
The molecular causes and mechanisms of neurodegenerative diseases remain poorly understood. A growing number of single-cell studies have implicated various neural, glial, and immune cell subtypes to affect the mammalian central nervous system in many age-related disorders. Integrating this body of transcriptomic evidence into a comprehensive and reproducible framework poses several computational challenges. Here, we introduce ZEBRA, a large single-cell and single-nucleus RNA-seq database. ZEBRA integrates and normalizes gene expression and metadata from 33 studies, encompassing 4.2 million human and mouse brain cells sampled from 39 brain regions. It incorporates samples from patients with neurodegenerative diseases like Alzheimer’s disease, Parkinson’s disease, and Multiple sclerosis, as well as samples from relevant mouse models. We employed scVI, a deep probabilistic auto-encoder model, to integrate the samples and curated both cell and sample metadata for downstream analysis. ZEBRA allows for cell-type and disease-specific markers to be explored and compared between sample conditions and brain regions, a cell composition analysis, and gene-wise feature mappings. Our comprehensive molecular database facilitates the generation of data-driven hypotheses, enhancing our understanding of mammalian brain function during aging and disease. The data sets, along with an interactive database are freely available at https://www.ccb.uni-saarland.de/zebra.
Graphical Abstract
Related collections
Most cited references60

- Record: found
- Abstract: found
- Article: found
NCBI GEO: archive for functional genomics data sets—update

- Record: found
- Abstract: found
- Article: found
Massively parallel digital transcriptional profiling of single cells
- Record: found
- Abstract: found
- Article: not found