- Record: found
- Abstract: found
- Article: found
Transcriptomic profiling to identify genes involved in Fusarium mycotoxin Deoxynivalenol and Zearalenone tolerance in the mycoparasitic fungus Clonostachys rosea
Read this article at
Abstract
Background
Clonostachys rosea strain IK726 is a mycoparasitic fungus capable of controlling mycotoxin-producing Fusarium species, including F. graminearum and F. culmorum, known to produce Zearalenone (ZEA) and Deoxynivalenol (DON). DON is a type B trichothecene known to interfere with protein synthesis in eukaryotes. ZEA is a estrogenic-mimicing mycotoxin that exhibits antifungal growth. C. rosea produces the enzyme zearalenone hydrolase (ZHD101), which degrades ZEA. However, the molecular basis of resistance to DON in C. rosea is not understood. We have exploited a genome-wide transcriptomic approach to identify genes induced by DON and ZEA in order to investigate the molecular basis of mycotoxin resistance C. rosea.
Results
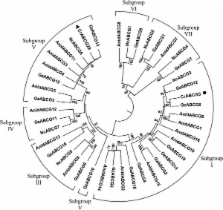
We generated DON- and ZEA-induced cDNA libraries based on suppression subtractive hybridization. A total of 443 and 446 sequenced clones (corresponding to 58 and 65 genes) from the DON- and ZEA-induced library, respectively, were analysed. DON-induced transcripts represented genes encoding metabolic enzymes such as cytochrome P450, cytochrome c oxidase and stress response proteins. In contrast, transcripts encoding the ZEA-detoxifying enzyme ZHD101 and those encoding a number of ATP-Binding Cassette (ABC) transporter transcripts were highly frequent in the ZEA-induced library. Subsequent bioinformatics analysis predicted that all transcripts with similarity to ABC transporters could be ascribed to only 2 ABC transporters genes, and phylogenetic analysis of the predicted ABC transporters suggested that they belong to group G (pleiotropic drug transporters) of the fungal ABC transporter gene family. This is the first report suggesting involvement of ABC transporters in ZEA tolerance. Expression patterns of a selected set of DON- and ZEA-induced genes were validated by the use of quantitative RT-PCR after exposure to the toxins. The qRT-PCR results obtained confirm the expression patterns suggested from the EST redundancy data.
Conclusion
The present study identifies a number of transcripts encoding proteins that are potentially involved in conferring resistance to DON and ZEA in the mycoparasitic fungus C. rosea. Whilst metabolic readjustment is potentially the key to withstanding DON, the fungus produces ZHD101 to detoxify ZEA and ABC transporters to transport ZEA or its degradation products out from the fungal cell.
Related collections
Most cited references39
- Record: found
- Abstract: not found
- Article: not found
The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins.
- Record: found
- Abstract: found
- Article: not found
Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: an oestrogenic mycotoxin.

- Record: found
- Abstract: found
- Article: found