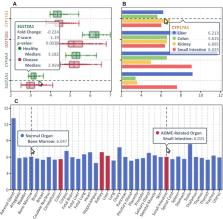
Journal ID (nlm-ta): Nucleic Acids Res
Journal ID (iso-abbrev): Nucleic Acids Res
Journal ID (publisher-id): nar
Title:
Nucleic Acids Research
Publisher:
Oxford University Press
ISSN
(Print):
0305-1048
ISSN
(Electronic):
1362-4962
Publication date Collection: 06
January
2023
Publication date
(Electronic):
16
October
2022
Publication date PMC-release: 16
October
2022
Volume: 51
Issue: D1
Pages: D1288-D1299
Affiliations
College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University
School of Medicine, Zhejiang University , Hangzhou 310058, China
Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University,
Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare , Hangzhou 330110, China
College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University
School of Medicine, Zhejiang University , Hangzhou 310058, China
College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University
School of Medicine, Zhejiang University , Hangzhou 310058, China
College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University
School of Medicine, Zhejiang University , Hangzhou 310058, China
Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University,
Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare , Hangzhou 330110, China
Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University,
Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare , Hangzhou 330110, China
State Key Laboratory of Chemical Oncogenomics, Key Laboratory of Chemical Biology,
Tsinghua Shenzhen International Graduate School, Tsinghua University , Shenzhen 518055, China
Qian Xuesen Collaborative Research Center of Astrochemistry and Space Life Sciences,
Institute of Drug Discovery Technology, Ningbo University , Ningbo 315211, China
Qian Xuesen Collaborative Research Center of Astrochemistry and Space Life Sciences,
Institute of Drug Discovery Technology, Ningbo University , Ningbo 315211, China
College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University
School of Medicine, Zhejiang University , Hangzhou 310058, China
College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University
School of Medicine, Zhejiang University , Hangzhou 310058, China
College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University
School of Medicine, Zhejiang University , Hangzhou 310058, China
State Key Laboratory of Chemical Oncogenomics, Key Laboratory of Chemical Biology,
Tsinghua Shenzhen International Graduate School, Tsinghua University , Shenzhen 518055, China
Qian Xuesen Collaborative Research Center of Astrochemistry and Space Life Sciences,
Institute of Drug Discovery Technology, Ningbo University , Ningbo 315211, China
College of Pharmaceutical Sciences, The Second Affiliated Hospital, Zhejiang University
School of Medicine, Zhejiang University , Hangzhou 310058, China
Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University,
Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare , Hangzhou 330110, China
Author notes
The authors wish it to be known that, in their opinion, the first two authors should
be regarded as Joint First Authors.
Author information
Article
Publisher ID:
gkac813
DOI: 10.1093/nar/gkac813
PMC ID: 9825453
PubMed ID: 36243961
SO-VID: f518ea1f-58b2-4125-afb6-ec273c1c1bb8
Copyright © © The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids
Research.
License:
This is an Open Access article distributed under the terms of the Creative Commons
Attribution-NonCommercial License (
https://creativecommons.org/licenses/by-nc/4.0/), which permits non-commercial re-use, distribution, and reproduction in any medium,
provided the original work is properly cited. For commercial re-use, please contact
journals.permissions@
123456oup.com
Funded by:
Natural Science Foundation of Zhejiang Province, DOI 10.13039/501100004731;
Award ID: LR21H300001
Funded by:
National Natural Science Foundation of China, DOI 10.13039/501100001809;
Award ID: 81872798
Award ID: U1909208
Funded by:
Leading Talent of the ‘Ten Thousand Plan’—National High-Level Talents Special Support
Plan of China;
Funded by:
Fundamental Research Fund of Central University;
Award ID: 2018QNA7023
Funded by:
Key R&D Program of Zhejiang Province;
Award ID: 2020C03010
Funded by:
National Key R&D Program of China Synthetic Biology Research;
Award ID: 2019YFA0905900
Funded by:
‘Double Top-Class’ University;
Award ID: 181201*194232101
Funded by:
Space Exploration Breeding Grant of Qian Xuesen Lab;
Award ID: TKTSPY-2020-04-03
Funded by:
Scientific Research Grant of Ningbo University;
Award ID: 215–432000282
Funded by:
Ningbo Top Talent Proj;
Award ID: 215–432094250
Funded by:
Shenzhen Municipal Government grant;
Award ID: JCYJ20170413113448742
Funded by:
Department of Science and Technology of Guangdong Province, DOI 10.13039/501100007162;
Award ID: 2017B030314083
Funded by:
Westlake Laboratory;
Funded by:
Alibaba–Zhejiang University Joint Research Center of Future Digital Healthcare;
Funded by:
Alibaba Cloud;
Funded by:
Information Technology Center of Zhejiang University;