- Record: found
- Abstract: found
- Article: found
Ensconsin/Map7 promotes microtubule growth and centrosome separation in Drosophila neural stem cells
Read this article at
Abstract
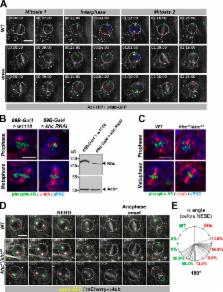
Ensconsin cooperates with its binding partner, Kinesin-1, during interphase to trigger centrosome separation, but it promotes microtubule polymerization independently of Kinesin-1 to control spindle length during mitosis.
Abstract
The mitotic spindle is crucial to achieve segregation of sister chromatids. To identify new mitotic spindle assembly regulators, we isolated 855 microtubule-associated proteins (MAPs) from Drosophila melanogaster mitotic or interphasic embryos. Using RNAi, we screened 96 poorly characterized genes in the Drosophila central nervous system to establish their possible role during spindle assembly. We found that Ensconsin/MAP7 mutant neuroblasts display shorter metaphase spindles, a defect caused by a reduced microtubule polymerization rate and enhanced by centrosome ablation. In agreement with a direct effect in regulating spindle length, Ensconsin overexpression triggered an increase in spindle length in S2 cells, whereas purified Ensconsin stimulated microtubule polymerization in vitro. Interestingly, ensc-null mutant flies also display defective centrosome separation and positioning during interphase, a phenotype also detected in kinesin-1 mutants. Collectively, our results suggest that Ensconsin cooperates with its binding partner Kinesin-1 during interphase to trigger centrosome separation. In addition, Ensconsin promotes microtubule polymerization during mitosis to control spindle length independent of Kinesin-1.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search.
- Record: found
- Abstract: found
- Article: not found
A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila.
- Record: found
- Abstract: found
- Article: not found