- Record: found
- Abstract: found
- Article: found
Rbfox1 Downregulation and Altered Calpain 3 Splicing by FRG1 in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
Read this article at
Abstract
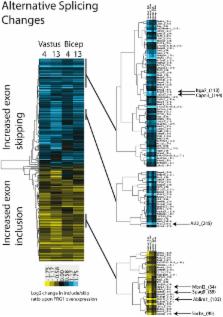
Facioscapulohumeral muscular dystrophy (FSHD) is a common muscle disease whose molecular pathogenesis remains largely unknown. Over-expression of FSHD region gene 1 ( FRG1) in mice, frogs, and worms perturbs muscle development and causes FSHD–like phenotypes. FRG1 has been implicated in splicing, and we asked how splicing might be involved in FSHD by conducting a genome-wide analysis in FRG1 mice. We find that splicing perturbations parallel the responses of different muscles to FRG1 over-expression and disease progression. Interestingly, binding sites for the Rbfox family of splicing factors are over-represented in a subset of FRG1-affected splicing events. Rbfox1 knockdown, over-expression, and RNA-IP confirm that these are direct Rbfox1 targets. We find that FRG1 is associated to the Rbfox1 RNA and decreases its stability. Consistent with this, Rbfox1 expression is down-regulated in mice and cells over-expressing FRG1 as well as in FSHD patients. Among the genes affected is Calpain 3, which is mutated in limb girdle muscular dystrophy, a disease phenotypically similar to FSHD. In FRG1 mice and FSHD patients, the Calpain 3 isoform lacking exon 6 ( Capn3 E6–) is increased. Finally, Rbfox1 knockdown and over-expression of Capn3 E6- inhibit muscle differentiation. Collectively, our results suggest that a component of FSHD pathogenesis may arise by over-expression of FRG1, reducing Rbfox1 levels and leading to aberrant expression of an altered Calpain 3 protein through dysregulated splicing.
Author Summary
Alternative splicing is a major contributor to the complexity of human cells, and its disruption can lead to a wide range of human disorders. FSHD is one of the most important muscle diseases. While muscle differentiation defects have been widely reported in the disease, the molecular mechanisms responsible are largely unknown. We found that expression of the alternative splicing factor Rbfox1 is a direct FRG1 target, and its expression decreased in the muscles of a mouse model of FSHD and FSHD patients. Moreover, alternative splicing of Calpain 3, encoding for a protease involved in muscle differentiation, is regulated by Rbfox1 and is altered in the muscles of the mouse model of FSHD and FSHD patients. Interestingly, we found that Rbfox1 is required for muscle differentiation and that this activity is likely mediated by Calpain 3 alternative splicing. Hence, our results suggest that decreased expression of Rbfox1 and aberrant Calpain 3 splicing contribute to the muscle differentiation defects of FSHD patients.
Related collections
Most cited references73
- Record: found
- Abstract: found
- Article: not found
Cluster analysis and display of genome-wide expression patterns.
- Record: found
- Abstract: found
- Article: not found
A unifying genetic model for facioscapulohumeral muscular dystrophy.
- Record: found
- Abstract: found
- Article: not found