- Record: found
- Abstract: found
- Article: found
Telomerase Is Essential for Zebrafish Heart Regeneration

Read this article at
SUMMARY
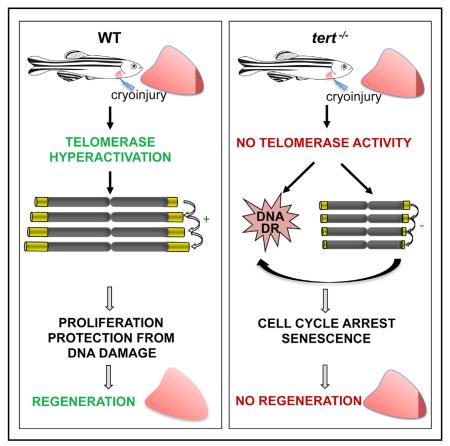
After myocardial infarction in humans, lost cardiomyocytes are replaced by an irreversible fibrotic scar. In contrast, zebrafish hearts efficiently regenerate after injury. Complete regeneration of the zebrafish heart is driven by the strong proliferation response of its cardiomyocytes to injury. Here we show that, after cardiac injury in zebrafish, telomerase becomes hyperactivated, and telomeres elongate transiently, preceding a peak of cardiomyocyte proliferation and full organ recovery. Using a telomerase-mutant zebrafish model, we found that telomerase loss drastically decreases cardiomyocyte proliferation and fibrotic tissue regression after cryoinjury and that cardiac function does not recover. The impaired cardiomyocyte proliferation response is accompanied by the absence of cardiomyocytes with long telomeres and an increased proportion of cardiomyocytes showing DNA damage and senescence characteristics. These findings demonstrate the importance of telomerase function in heart regeneration and highlight the potential of telomerase therapy as a means of stimulating cell proliferation upon myocardial infarction.
Graphical Abstract
In Brief
Bednarek et al. find that telomerase, well known for its role in elongating telomere ends, is essential during zebrafish heart regeneration. Cardiac injury hyperactivates telomerase and increases telomere length in cardiac cells. In telomerase-null mutants, cardiac cells accumulate DNA damage and do not efficiently proliferate in response to injury.
Related collections
Most cited references37
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found
The inflammatory response in myocardial injury, repair, and remodelling.
- Record: found
- Abstract: found
- Article: not found