- Record: found
- Abstract: found
- Article: found
Cross‐species comparison illuminates the importance of iron homeostasis for splenic anti‐immunosenescence
Read this article at
Abstract
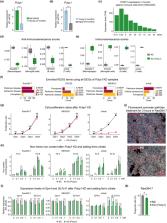
Although immunosenescence may result in increased morbidity and mortality, many mammals have evolved effective immune coping strategies to extend their lifespans. Thus, the immune systems of long‐lived mammals present unique models to study healthy longevity. To identify the molecular clues of anti‐immunosenescence, we first built high‐quality reference genome for a long‐lived myotis bat, and then compared three long‐lived mammals (i.e., bat, naked mole rat, and human) versus the short‐lived mammal, mouse, in splenic immune cells at single‐cell resolution. A close relationship between B:T cell ratio and immunosenescence was detected, as B:T cell ratio was much higher in mouse than long‐lived mammals and significantly increased during aging. Importantly, we identified several iron‐related genes that could resist immunosenescence changes, especially the iron chaperon, PCBP1, which was upregulated in long‐lived mammals but dramatically downregulated during aging in all splenic immune cell types. Supportively, immune cells of mouse spleens contained more free iron than those of bat spleens, suggesting higher level of ROS‐induced damage in mouse. PCBP1 downregulation during aging was also detected in hepatic but not pulmonary immune cells, which is consistent with the crucial roles of spleen and liver in organismal iron recycling. Furthermore, PCBP1 perturbation in immune cell lines would result in cellular iron dyshomeostasis and senescence. Finally, we identified two transcription factors that could regulate PCBP1 during aging. Together, our findings highlight the importance of iron homeostasis in splenic anti‐immunosenescence, and provide unique insight for improving human healthspan.
Abstract
To identify molecular clues of anti‐immunosenescence, we compare three long‐lived mammals versus the short‐lived mammal, mouse, in splenic immune cells at single‐cell resolution. We detect higher splenic B:T cell ratio and higher free iron level in mice, while higher expression levels of iron homeostasis‐related genes in long‐lived species. Our findings highlight the importance of iron homeostasis in splenic anti‐immunosenescence, and provide unique insight for improving human healthspan.
Related collections
Most cited references58

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.

- Record: found
- Abstract: found
- Article: found
