- Record: found
- Abstract: found
- Article: found
Alu insertion variants alter mRNA splicing
Read this article at
Abstract
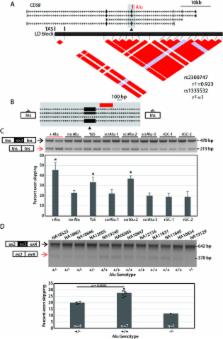
RNA splicing is a highly regulated process dependent on sequences near splice sites. Insertions of Alu retrotransposons can disrupt splice sites or bind splicing regulators. We hypothesized that some common inherited polymorphic Alu insertions are responsible for splicing QTLs (sQTL). We focused on intronic Alu variants mapping within 100 bp of an alternatively used exon and screened for those that alter splicing. We identify five loci, 21.7% of those assayed, where the polymorphic Alu alters splicing. While in most cases the Alu promotes exon skipping, at one locus the Alu increases exon inclusion. Of particular interest is an Alu polymorphism in the CD58 gene. Reduced CD58 expression is associated with risk for developing multiple sclerosis. We show that the Alu insertion promotes skipping of CD58 exon 3 and results in a frameshifted transcript, indicating that the Alu may be the causative variant for increased MS risk at this locus. Using RT-PCR analysis at the endogenous locus, we confirm that the Alu variant is a sQTL for CD58. In summary, altered splicing efficiency is a common functional consequence of Alu polymorphisms including at least one instance where the variant is implicated in disease risk. This work broadens our understanding of splicing regulatory sequences around exons.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
Epithelial-mesenchymal transitions in development and disease.
- Record: found
- Abstract: found
- Article: not found
Understanding alternative splicing: towards a cellular code.
- Record: found
- Abstract: not found
- Article: not found