- Record: found
- Abstract: found
- Article: not found
Measurements of translation initiation from all 64 codons in E. coli
Read this article at
Abstract
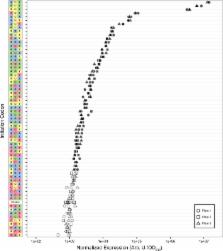
Our understanding of translation underpins our capacity to engineer living systems. The canonical start codon (AUG) and a few near-cognates (GUG, UUG) are considered as the ‘start codons’ for translation initiation in Escherichia coli. Translation is typically not thought to initiate from the 61 remaining codons. Here, we quantified translation initiation of green fluorescent protein and nanoluciferase in E. coli from all 64 triplet codons and across a range of DNA copy number. We detected initiation of protein synthesis above measurement background for 47 codons. Translation from non-canonical start codons ranged from 0.007 to 3% relative to translation from AUG. Translation from 17 non-AUG codons exceeded the highest reported rates of non-cognate codon recognition. Translation initiation from non-canonical start codons may contribute to the synthesis of peptides in both natural and synthetic biological systems.
Related collections
Most cited references115
- Record: found
- Abstract: not found
- Article: not found
Enzymatic assembly of DNA molecules up to several hundred kilobases.
- Record: found
- Abstract: found
- Article: not found
Simultaneous inference in general parametric models.
- Record: found
- Abstract: found
- Article: not found
Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling.
Author and article information
Comments
Comment on this article
Similar content166
- La traducción al español de textos jurídicos-institucionales franceses: principales parámetros orientados a los alumnos de Traducción e Interpretación Translated title: Translation into Spanish of French juridical-institutional texts: main parameters orientated to Translation and Interpretation studentsAuthors: José María Castellano Martínez
