- Record: found
- Abstract: found
- Article: not found
MaizeGDB update: new tools, data and interface for the maize model organism database
Read this article at
Abstract
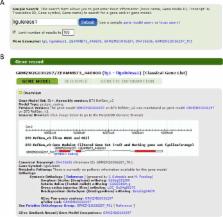
MaizeGDB is a highly curated, community-oriented database and informatics service to researchers focused on the crop plant and model organism Zea mays ssp. mays. Although some form of the maize community database has existed over the last 25 years, there have only been two major releases. In 1991, the original maize genetics database MaizeDB was created. In 2003, the combined contents of MaizeDB and the sequence data from ZmDB were made accessible as a single resource named MaizeGDB. Over the next decade, MaizeGDB became more sequence driven while still maintaining traditional maize genetics datasets. This enabled the project to meet the continued growing and evolving needs of the maize research community, yet the interface and underlying infrastructure remained unchanged. In 2015, the MaizeGDB team completed a multi-year effort to update the MaizeGDB resource by reorganizing existing data, upgrading hardware and infrastructure, creating new tools, incorporating new data types (including diversity data, expression data, gene models, and metabolic pathways), and developing and deploying a modern interface. In addition to coordinating a data resource, the MaizeGDB team coordinates activities and provides technical support to the maize research community. MaizeGDB is accessible online at http://www.maizegdb.org.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
The generic genome browser: a building block for a model organism system database.
- Record: found
- Abstract: found
- Article: not found
