- Record: found
- Abstract: found
- Article: found
Breast Cancer DNA Methylation Profiles Are Associated with Tumor Size and Alcohol and Folate Intake
Read this article at
Abstract
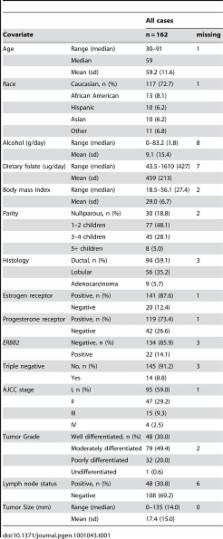
Although tumor size and lymph node involvement are the current cornerstones of breast cancer prognosis, they have not been extensively explored in relation to tumor methylation attributes in conjunction with other tumor and patient dietary and hormonal characteristics. Using primary breast tumors from 162 (AJCC stage I–IV) women from the Kaiser Division of Research Pathways Study and the Illumina GoldenGate methylation bead-array platform, we measured 1,413 autosomal CpG loci associated with 773 cancer-related genes and validated select CpG loci with Sequenom EpiTYPER. Tumor grade, size, estrogen and progesterone receptor status, and triple negative status were significantly ( Q-values <0.05) associated with altered methylation of 209, 74, 183, 69, and 130 loci, respectively. Unsupervised clustering, using a recursively partitioned mixture model (RPMM), of all autosomal CpG loci revealed eight distinct methylation classes. Methylation class membership was significantly associated with patient race ( P<0.02) and tumor size ( P<0.001) in univariate tests. Using multinomial logistic regression to adjust for potential confounders, patient age and tumor size, as well as known disease risk factors of alcohol intake and total dietary folate, were all significantly ( P<0.0001) associated with methylation class membership. Breast cancer prognostic characteristics and risk-related exposures appear to be associated with gene-specific tumor methylation, as well as overall methylation patterns.
Author Summary
The current standard prognostic indicator for breast cancer is tumor-node-metastasis staging; though, as population-based studies and clinical trials are conducted, molecular characterization of disease is beginning to allow improved markers of prognosis and assist clinicians in choosing the most appropriate therapies. We investigated DNA methylation profiles in over 160 well annotated breast tumor samples and found significant relationships with standard and other known predictors of prognosis, as well as established risk factors for disease: alcohol intake and dietary folate. Recently the United States National Cancer Institute Cancer Biomarkers Research Group articulated a need for a “Strategic Approach to Validating Methylated Genes as Biomarkers for Breast Cancer,” and our work is extremely responsive to this call for a national strategy. Recognizing the increasing use of pre-operative chemotherapy for patients with operable, early-stage disease, there is added complexity in breast cancer staging. Since chemotherapy can considerably decrease tumor size, it is still unclear whether pre-operative or post-operative stage best informs prognosis and treatment decisions for patients electing pre-operative chemotherapy. However, our data clearly illustrate the promise of tumor DNA methylation for augmenting tumor staging and can be attained with minimal tissue in a pre-operative context.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Effect of preoperative chemotherapy on the outcome of women with operable breast cancer.
- Record: found
- Abstract: found
- Article: not found
Effect of preoperative chemotherapy on local-regional disease in women with operable breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B-18.
- Record: found
- Abstract: found
- Article: not found