- Record: found
- Abstract: found
- Article: found
Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
Read this article at
Abstract
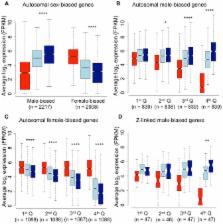
Gene expression differences between the sexes account for the majority of sexually dimorphic phenotypes, and the study of sex-biased gene expression is important for understanding the genetic basis of complex sexual dimorphisms. However, it has been difficult to test the nature of this relationship due to the fact that sexual dimorphism has traditionally been conceptualized as a dichotomy between males and females, rather than an axis with individuals distributed at intermediate points. The wild turkey ( Meleagris gallopavo) exhibits just this sort of continuum, with dominant and subordinate males forming a gradient in male secondary sexual characteristics. This makes it possible for the first time to test the correlation between sex-biased gene expression and sexually dimorphic phenotypes, a relationship crucial to molecular studies of sexual selection and sexual conflict. Here, we show that subordinate male transcriptomes show striking multiple concordances with their relative phenotypic sexual dimorphism. Subordinate males were clearly male rather than intersex, and when compared to dominant males, their transcriptomes were simultaneously demasculinized for male-biased genes and feminized for female-biased genes across the majority of the transcriptome. These results provide the first evidence linking sexually dimorphic transcription and sexually dimorphic phenotypes. More importantly, they indicate that evolutionary changes in sexual dimorphism can be achieved by varying the magnitude of sex-bias in expression across a large proportion of the coding content of a genome.
Author Summary
Males and females exhibit many differences in morphology, behavior and physiology, yet they share the vast majority of their genomes. Most differences between the sexes are therefore thought to be the product of gene expression differences between females and males. Studies of sex differences in expression assume that genes expressed more in males encode male traits, and genes expressed more in females encode female traits, and this assumption is a key foundation to genetic studies of sexual dimorphism and sexual conflict. Despite this key assumption, this relationship has yet to be empirically tested, as the main model organisms for studies of sex-biased gene expression lack multiple male and female morphs. Here, we use the two male morphs in the wild turkey to show that the magnitude of male-biased gene expression correlates with the manifestation of sexually dimorphic traits. Males with less manifestation of sexual dimorphism in phenotype were both demasculinized for male-biased genes, as well as feminized for female-biased genes. This pattern encompassed the majority of expressed loci, suggesting that evolutionary changes in the magnitude of sexual dimorphism may be achieved by small changes in the magnitude of sex-biased transcription across thousands of genes.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
Ensembl 2012
- Record: found
- Abstract: found
- Article: not found
Sex-dependent gene expression and evolution of the Drosophila transcriptome.
- Record: found
- Abstract: found
- Article: not found