- Record: found
- Abstract: found
- Article: found
Support for Lungfish as the Closest Relative of Tetrapods by Using Slowly Evolving Ray-Finned Fish as the Outgroup
Read this article at
Abstract
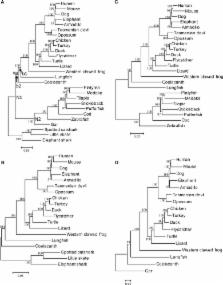
In a previous analysis of the phylogenetic relationships of coelacanths, lungfishes and tetrapods, using cartilaginous fish (CF) as the outgroup, the sister relationship of lungfishes and tetrapods was constructed with high statistical support. However, using as the outgroup ray-finned fish (RF), which are more taxonomically closely related to the three lineages than CF, the sister relationship of coelacanths and tetrapods was most often constructed depending on the methods and the data sets, but the statistical support was generally low except in the cases in which the data set including a small number of species was analyzed. In this study, instead of the fast evolving ray-finned fish, teleost fish (TF), in the previous data sets, by using two slowly evolving RF, gar and bowfin, as the outgroup, we showed that the sister relationship of lungfishes and tetrapods was reconstructed with high statistical support. In our analysis the evolutionary rates of gar and bowfin were similar to each other and one third to one half of TF. The difference of the amino acid frequencies of the two species with other lineages was larger than those of TF. This study provides a strong support for lungfishes as the closest relative of tetrapods and indicates the importance of using an appropriate outgroup with small divergence in phylogenetic construction.
Related collections
Most cited references38

- Record: found
- Abstract: found
- Article: found
ASTRAL: genome-scale coalescent-based species tree estimation

- Record: found
- Abstract: found
- Article: found
Elephant shark genome provides unique insights into gnathostome evolution
- Record: found
- Abstract: found
- Article: not found