- Record: found
- Abstract: found
- Article: found
A Functional Switch of NuRD Chromatin Remodeling Complex Subunits Regulates Mouse Cortical Development
Read this article at
Summary
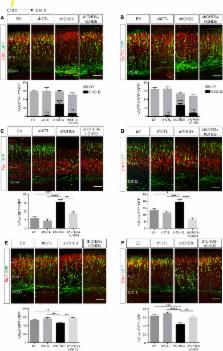
Histone modifications and chromatin remodeling represent universal mechanisms by which cells adapt their transcriptional response to rapidly changing environmental conditions. Extensive chromatin remodeling takes place during neuronal development, allowing the transition of pluripotent cells into differentiated neurons. Here, we report that the NuRD complex, which couples ATP-dependent chromatin remodeling with histone deacetylase activity, regulates mouse brain development. Subunit exchange of CHDs, the core ATPase subunits of the NuRD complex, is required for distinct aspects of cortical development. Whereas CHD4 promotes the early proliferation of progenitors, CHD5 facilitates neuronal migration and CHD3 ensures proper layer specification. Inhibition of each CHD leads to defects of neuronal differentiation and migration, which cannot be rescued by expressing heterologous CHDs. Finally, we demonstrate that NuRD complexes containing specific CHDs are recruited to regulatory elements and modulate the expression of genes essential for brain development.
Graphical Abstract
Highlights
-
•
The ATPases CHD3, CHD4, and CHD5 are mutually exclusive subunits of the NuRD complex
-
•
CHD3, CHD4, and CHD5 regulate distinct and non-redundant aspects of cortical development
-
•
Loss of each CHD leads to specific defects of neuronal proliferation and migration
-
•
CHD3, CHD4, and CHD5 regulate distinct set of genes essential for brain development
Abstract
Neural development requires active chromatin remodeling. Nitarska et al. identify distinct NuRD chromatin remodeling complexes, containing CHD3, CHD4, or CHD5, during mouse embryonic cortical development. CHD4 promotes proliferation of basal progenitors, while CHD5 facilitates early radial migration and CHD3 drives late migration and laminar specification of neurons.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo.
- Record: found
- Abstract: found
- Article: not found
Neural progenitors, neurogenesis and the evolution of the neocortex.
- Record: found
- Abstract: found
- Article: not found
