- Record: found
- Abstract: found
- Article: not found
Ribosomal genes in focus : new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ
Read this article at
Abstract
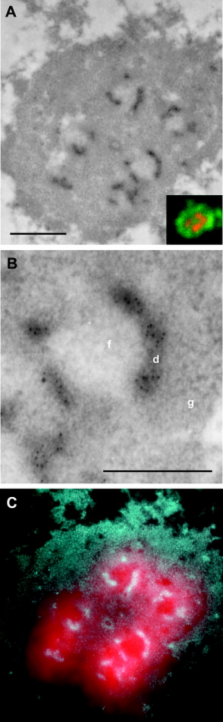
T he organization of transcriptionally active ribosomal genes in animal cell nucleoli is investigated in this study in order to address the long-standing controversy with regard to the intranucleolar localization of these genes. Detailed analyses of HeLa cell nucleoli include direct localization of ribosomal genes by in situ hybridization and their indirect localization via nascent ribosomal transcript mappings. On the light microscopy (LM) level, ribosomal genes map in 10–40 fluorescence foci per nucleus, and transcription activity is associated with most foci. We demonstrate that each nucleolar focus observed by LM corresponds, on the EM level, to an individual fibrillar center (FC) and surrounding dense fibrillar components (DFCs). The EM data identify the DFC as the nucleolar subcompartment in which rRNA synthesis takes place, consistent with detection of rDNA within the DFC. The highly sensitive method for mapping nascent transcripts in permeabilized cells on ultrastructural level provides intense and unambiguous clustered immunogold signal over the DFC, whereas very little to no label is detected over the FC. This signal is strongly indicative of nascent “Christmas trees” of rRNA associated with individual rDNA genes, sampled on the surface of thin sections. Stereological analysis of the clustered transcription signal further suggests that these Christmas trees may be contorted in space and exhibit a DNA compaction ratio on the order of 4–5.5.
Related collections
Most cited references17
- Record: found
- Abstract: found
- Article: not found
Visualization of focal sites of transcription within human nuclei.
- Record: found
- Abstract: found
- Article: not found
Visualization of nucleolar genes.
- Record: found
- Abstract: found
- Article: not found
