- Record: found
- Abstract: found
- Article: found
Transposable Elements: Powerful Contributors to Angiosperm Evolution and Diversity
Read this article at
Abstract
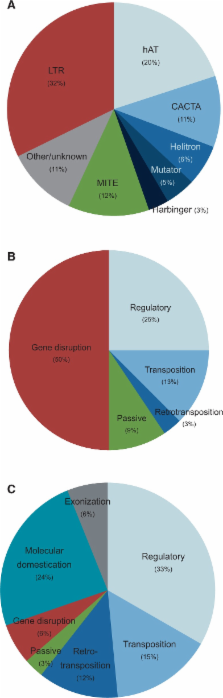
Transposable elements (TEs) are a dominant feature of most flowering plant genomes. Together with other accepted facilitators of evolution, accumulating data indicate that TEs can explain much about their rapid evolution and diversification. Genome size in angiosperms is highly correlated with TE content and the overwhelming bulk (>80%) of large genomes can be composed of TEs. Among retro-TEs, long terminal repeats (LTRs) are abundant, whereas DNA-TEs, which are often less abundant than retro-TEs, are more active. Much adaptive or evolutionary potential in angiosperms is due to the activity of TEs (active TE-Thrust), resulting in an extraordinary array of genetic changes, including gene modifications, duplications, altered expression patterns, and exaptation to create novel genes, with occasional gene disruption. TEs implicated in the earliest origins of the angiosperms include the exapted Mustang, Sleeper, and Fhy3/Far1 gene families. Passive TE-Thrust can create a high degree of adaptive or evolutionary potential by engendering ectopic recombination events resulting in deletions, duplications, and karyotypic changes. TE activity can also alter epigenetic patterning, including that governing endosperm development, thus promoting reproductive isolation. Continuing evolution of long-lived resprouter angiosperms, together with genetic variation in their multiple meristems, indicates that TEs can facilitate somatic evolution in addition to germ line evolution. Critical to their success, angiosperms have a high frequency of polyploidy and hybridization, with resultant increased TE activity and introgression, and beneficial gene duplication. Together with traditional explanations, the enhanced genomic plasticity facilitated by TE-Thrust, suggests a more complete and satisfactory explanation for Darwin’s “abominable mystery”: the spectacular success of the angiosperms.
Related collections
Most cited references171
- Record: found
- Abstract: found
- Article: not found
The Sorghum bicolor genome and the diversification of grasses.
- Record: found
- Abstract: found
- Article: not found
Mobile elements: drivers of genome evolution.
- Record: found
- Abstract: found
- Article: not found