- Record: found
- Abstract: found
- Article: found
Ras GTPase-Like Protein MglA, a Controller of Bacterial Social-Motility in Myxobacteria, Has Evolved to Control Bacterial Predation by Bdellovibrio
Read this article at
Abstract
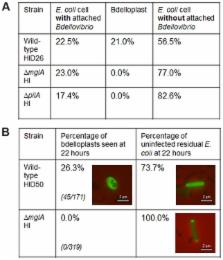
Bdellovibrio bacteriovorus invade Gram-negative bacteria in a predatory process requiring Type IV pili (T4P) at a single invasive pole, and also glide on surfaces to locate prey. Ras-like G-protein MglA, working with MglB and RomR in the deltaproteobacterium Myxococcus xanthus, regulates adventurous gliding and T4P-mediated social motility at both M. xanthus cell poles. Our bioinformatic analyses suggested that the GTPase activating protein (GAP)-encoding gene mglB was lost in Bdellovibrio, but critical residues for MglA Bd GTP-binding are conserved. Deletion of mglA Bd abolished prey-invasion, but not gliding, and reduced T4P formation. MglA Bd interacted with a previously uncharacterised tetratricopeptide repeat (TPR) domain protein Bd2492, which we show localises at the single invasive pole and is required for predation. Bd2492 and RomR also interacted with cyclic-di-GMP-binding receptor CdgA, required for rapid prey-invasion. Bd2492, RomR Bd and CdgA localize to the invasive pole and may facilitate MglA-docking. Bd2492 was encoded from an operon encoding a TamAB-like secretion system. The TamA protein and RomR were found, by gene deletion tests, to be essential for viability in both predatory and non-predatory modes. Control proteins, which regulate bipolar T4P-mediated social motility in swarming groups of deltaproteobacteria, have adapted in evolution to regulate the anti-social process of unipolar prey-invasion in the “lone-hunter” Bdellovibrio. Thus GTP-binding proteins and cyclic-di-GMP inputs combine at a regulatory hub, turning on prey-invasion and allowing invasion and killing of bacterial pathogens and consequent predatory growth of Bdellovibrio.
Author Summary
Bacterial cell polarity control is important for maintaining asymmetry of polar components such as flagella and pili. Bdellovibrio bacteriovorus is a predatory deltaproteobacterium which attaches to, and invades, other bacteria using Type IV pili (T4P) extruded from the specialised, invasive, non-flagellar pole of the cell. It was not known how that invasive pole is specified and regulated. Here we discover that a regulatory protein-hub, including Ras-GTPase-like protein MglA and cyclic-di-GMP receptor-protein CdgA, control prey-invasion. In the deltaproteobacterium, Myxococcus xanthus, MglA, with MglB and RomR, was found by others to regulate switching of T4P in social ‘swarming’ surface motility by swapping the pole at which T4P are found. In contrast, in B. bacteriovorus MglA regulates the process of prey-invasion and RomR, which is required for surface motility regulation in Myxococcus, is essential for growth and viability in Bdellovibrio. During evolution, B. bacteriovorus has lost mglB, possibly as T4P-pole-switching is not required; pili are only required at the invasive pole. A previously unidentified tetratricopeptide repeat (TPR) protein interacts with MglA and is essential for prey-invasion. This regulatory protein hub allows prey-invasion, likely integrating cyclic-di-GMP signals, pilus assembly and TamAB secretion in B. bacteriovorus.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli.
- Record: found
- Abstract: found
- Article: not found
Discovery of an archetypal protein transport system in bacterial outer membranes.
- Record: found
- Abstract: found
- Article: not found