- Record: found
- Abstract: found
- Article: found
Toward the sequence-based breeding in legumes in the post-genome sequencing era
Read this article at
Abstract
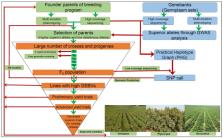
Efficiency of breeding programs of legume crops such as chickpea, pigeonpea and groundnut has been considerably improved over the past decade through deployment of modern genomic tools and technologies. For instance, next-generation sequencing technologies have facilitated availability of genome sequence assemblies, re-sequencing of several hundred lines, development of HapMaps, high-density genetic maps, a range of marker genotyping platforms and identification of markers associated with a number of agronomic traits in these legume crops. Although marker-assisted backcrossing and marker-assisted selection approaches have been used to develop superior lines in several cases, it is the need of the hour for continuous population improvement after every breeding cycle to accelerate genetic gain in the breeding programs. In this context, we propose a sequence-based breeding approach which includes use of independent or combination of parental selection, enhancing genetic diversity of breeding programs, forward breeding for early generation selection, and genomic selection using sequencing/genotyping technologies. Also, adoption of speed breeding technology by generating 4–6 generations per year will be contributing to accelerate genetic gain. While we see a huge potential of the sequence-based breeding to revolutionize crop improvement programs in these legumes, we anticipate several challenges especially associated with high-quality and precise phenotyping at affordable costs, data analysis and management related to improving breeding operation efficiency. Finally, integration of improved seed systems and better agronomic packages with the development of improved varieties by using sequence-based breeding will ensure higher genetic gains in farmers’ fields.
Related collections
Most cited references73
- Record: found
- Abstract: found
- Article: not found
QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations.
- Record: found
- Abstract: found
- Article: not found
High-throughput genotyping by whole-genome resequencing.
- Record: found
- Abstract: found
- Article: not found