- Record: found
- Abstract: found
- Article: found
Comparative population genomics reveals the domestication history of the peach, Prunus persica , and human influences on perennial fruit crops
Read this article at
Abstract
Background
Recently, many studies utilizing next generation sequencing have investigated plant evolution and domestication in annual crops. Peach, Prunus persica, is a typical perennial fruit crop that has ornamental and edible varieties. Unlike other fruit crops, cultivated peach includes a large number of phenotypes but few polymorphisms. In this study, we explore the genetic basis of domestication in peach and the influence of humans on its evolution.
Results
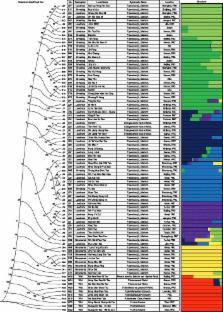
We perform large-scale resequencing of 10 wild and 74 cultivated peach varieties, including 9 ornamental, 23 breeding, and 42 landrace lines. We identify 4.6 million SNPs, a large number of which could explain the phenotypic variation in cultivated peach. Population analysis shows a single domestication event, the speciation of P. persica from wild peach. Ornamental and edible peach both belong to P. persica , along with another geographically separated subgroup, Prunus ferganensis.
We identify 147 and 262 genes under edible and ornamental selection, respectively. Some of these genes are associated with important biological features. We perform a population heterozygosity analysis in different plants that indicates that free recombination effects could affect domestication history. By applying artificial selection during the domestication of the peach and facilitating its asexual propagation, humans have caused a sharp decline of the heterozygote ratio of SNPs.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla.
- Record: found
- Abstract: found
- Article: not found
Rapid isolation of high molecular weight plant DNA.
- Record: found
- Abstract: found
- Article: not found