- Record: found
- Abstract: found
- Article: found
A Point Mutation in Suppressor of Cytokine Signalling 2 ( Socs2) Increases the Susceptibility to Inflammation of the Mammary Gland while Associated with Higher Body Weight and Size and Higher Milk Production in a Sheep Model
Read this article at
Abstract
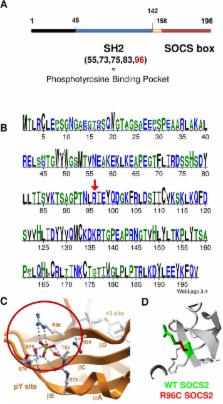
Mastitis is an infectious disease mainly caused by bacteria invading the mammary gland. Genetic control of susceptibility to mastitis has been widely evidenced in dairy ruminants, but the genetic basis and underlying mechanisms are still largely unknown. We describe the discovery, fine mapping and functional characterization of a genetic variant associated with elevated milk leukocytes count, or SCC, as a proxy for mastitis. After implementing genome-wide association studies, we identified a major QTL associated with SCC on ovine chromosome 3. Fine mapping of the region, using full sequencing with 12X coverage in three animals, provided one strong candidate SNP that mapped to the coding sequence of a highly conserved gene, suppressor of cytokine signalling 2 ( Socs2). The frequency of the SNP associated with increased SCC was 21.7% and the Socs2 genotype explained 12% of the variance of the trait. The point mutation induces the p.R96C substitution in the SH2 functional domain of SOCS2 i.e. the binding site of the protein to various ligands, as well-established for the growth hormone receptor GHR. Using surface plasmon resonance we showed that the p.R96C point mutation completely abrogates SOCS2 binding affinity for the phosphopeptide of GHR. Additionally, the size, weight and milk production in p.R96C homozygote sheep, were significantly increased by 24%, 18%, and 4.4%, respectively, when compared to wild type sheep, supporting the view that the point mutation causes a loss of SOCS2 functional activity. Altogether these results provide strong evidence for a causal mutation controlling SCC in sheep and highlight the major role of SOCS2 as a tradeoff between the host’s inflammatory response to mammary infections, and body growth and milk production, which are all mediated by the JAK/STAT signaling pathway.
Author Summary
Mastitis is an inflammation of the mammary gland mainly caused by invading bacteria. Ruminants show natural variability in their predisposition to mastitis, and therefore provide unique models for study of the genetics and physiology of host response to bacterial infection. A genome-wide association study was conducted in a dairy sheep population for milk somatic cell counts as a proxy for mastitis. Fine mapping, using whole genome sequencing, led to the identification of a mutation in the Suppressor of Cytokine Signaling 2 gene (socs2). This mutation was shown to cause a loss of functional activity of the SOCS2 protein, which suggested impairment of feedback control of the JAK/STAT signaling pathways in susceptible animals. Additionally, size, weight and milk production were increased in animals carrying the susceptible variant suggesting a pleiotropic effect of the gene on production versus health traits. Results gave strong evidence of the role of SOCS2 in the host’s inflammation of the udder and provided new insights into the key mechanisms underlying the genetic control of mastitis.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
The genome sequence of taurine cattle: a window to ruminant biology and evolution.
- Record: found
- Abstract: found
- Article: not found
Mapping mendelian factors underlying quantitative traits using RFLP linkage maps.
- Record: found
- Abstract: found
- Article: not found