- Record: found
- Abstract: found
- Article: found
Gene Therapy for Cystic Fibrosis: Progress and Challenges of Genome Editing
Read this article at
Abstract
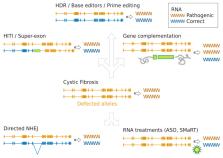
Since the early days of its conceptualization and application, human gene transfer held the promise of a permanent solution to genetic diseases including cystic fibrosis (CF). This field went through alternated periods of enthusiasm and distrust. The development of refined technologies allowing site specific modification with programmable nucleases highly revived the gene therapy field. CRISPR nucleases and derived technologies tremendously facilitate genome manipulation offering diversified strategies to reverse mutations. Here we discuss the advancement of gene therapy, from therapeutic nucleic acids to genome editing techniques, designed to reverse genetic defects in CF. We provide a roadmap through technologies and strategies tailored to correct different types of mutations in the cystic fibrosis transmembrane regulator ( CFTR) gene, and their applications for the development of experimental models valuable for the advancement of CF therapies.
Related collections
Most cited references60
- Record: found
- Abstract: found
- Article: not found
Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements
- Record: found
- Abstract: found
- Article: not found
Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients.
- Record: found
- Abstract: found
- Article: not found