- Record: found
- Abstract: found
- Article: found
Weighted gene co-expression network analysis of expression data of monozygotic twins identifies specific modules and hub genes related to BMI
Read this article at
Abstract
Background
The therapeutic management of obesity is challenging, hence further elucidating the underlying mechanisms of obesity development and identifying new diagnostic biomarkers and therapeutic targets are urgent and necessary. Here, we performed differential gene expression analysis and weighted gene co-expression network analysis (WGCNA) to identify significant genes and specific modules related to BMI based on gene expression profile data of 7 discordant monozygotic twins.
Results
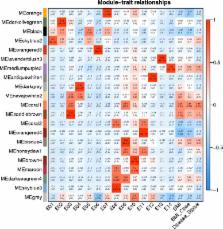
In the differential gene expression analysis, it appeared that 32 differentially expressed genes (DEGs) were with a trend of up-regulation in twins with higher BMI when compared to their siblings. Categories of positive regulation of nitric-oxide synthase biosynthetic process, positive regulation of NF-kappa B import into nucleus, and peroxidase activity were significantly enriched within GO database and NF-kappa B signaling pathway within KEGG database. DEGs of NAMPT, TLR9, PTGS2, HBD, and PCSK1N might be associated with obesity. In the WGCNA, among the total 20 distinct co-expression modules identified, coral1 module (68 genes) had the strongest positive correlation with BMI ( r = 0.56, P = 0.04) and disease status (r = 0.56, P = 0.04). Categories of positive regulation of phospholipase activity, high-density lipoprotein particle clearance, chylomicron remnant clearance, reverse cholesterol transport, intermediate-density lipoprotein particle, chylomicron, low-density lipoprotein particle, very-low-density lipoprotein particle, voltage-gated potassium channel complex, cholesterol transporter activity, and neuropeptide hormone activity were significantly enriched within GO database for this module. And alcoholism and cell adhesion molecules pathways were significantly enriched within KEGG database. Several hub genes, such as GAL, ASB9, NPPB, TBX2, IL17C, APOE, ABCG4, and APOC2 were also identified. The module eigengene of saddlebrown module (212 genes) was also significantly correlated with BMI ( r = 0.56, P = 0.04), and hub genes of KCNN1 and AQP10 were differentially expressed.
Conclusion
We identified significant genes and specific modules potentially related to BMI based on the gene expression profile data of monozygotic twins. The findings may help further elucidate the underlying mechanisms of obesity development and provide novel insights to research potential gene biomarkers and signaling pathways for obesity treatment. Further analysis and validation of the findings reported here are important and necessary when more sample size is acquired.
Related collections
Most cited references81
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: found