- Record: found
- Abstract: found
- Article: found
FlyBase at 25: looking to the future
Read this article at
Abstract
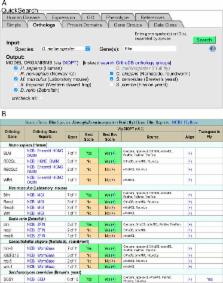
Since 1992, FlyBase ( flybase.org) has been an essential online resource for the Drosophila research community. Concentrating on the most extensively studied species, Drosophila melanogaster, FlyBase includes information on genes (molecular and genetic), transgenic constructs, phenotypes, genetic and physical interactions, and reagents such as stocks and cDNAs. Access to data is provided through a number of tools, reports, and bulk-data downloads. Looking to the future, FlyBase is expanding its focus to serve a broader scientific community. In this update, we describe new features, datasets, reagent collections, and data presentations that address this goal, including enhanced orthology data, Human Disease Model Reports, protein domain search and visualization, concise gene summaries, a portal for external resources, video tutorials and the FlyBase Community Advisory Group.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
Identification of functional elements and regulatory circuits by Drosophila modENCODE.
- Record: found
- Abstract: found
- Article: not found
Versatile P(acman) BAC Libraries for Transgenesis Studies in Drosophila melanogaster
- Record: found
- Abstract: found
- Article: not found