- Record: found
- Abstract: found
- Article: found
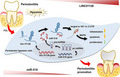
Long non‐coding RNA 01126 promotes periodontitis pathogenesis of human periodontal ligament cells via miR‐518a‐5p/HIF‐1α/MAPK pathway
Read this article at
Abstract
Background
Periodontitis is a prevalent oral inflammatory disease, which can cause periodontal ligament to a local hypoxia environment. However, the mechanism of hypoxia associated long non‐coding RNAs (lncRNAs) involved in periodontitis is still largely unknown.
Methods
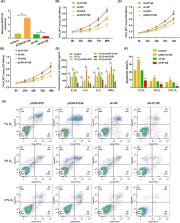
Microarray was performed to detect the expression patterns of lncRNAs in 3 pairs of gingival tissues from patients with periodontitis and healthy controls. The expression of lncRNA 01126 (LINC01126), miR‐518a‐5p and hypoxia‐inducible factor‐1α (HIF‐1α) in periodontal tissues and in human periodontal ligament cells (hPDLCs) under hypoxia was measured by quantitative real‐time polymerase chain reaction or western blot. Fluorescence in situ hybridization and cell fraction assay were performed to determine the subcellular localization of LINC01126 and miR‐518a‐5p. Overexpression or knockdown of LINC01126 or HIF‐1α was used to confirm their biological roles in hPDLCs. MTT assays were performed to evaluate hPDLCs proliferation ability. Flow cytometry was used to detect apoptosis. ELISA was used to measure the expression levels of interleukin (IL)‐1β, IL‐6, IL‐8 and TNF‐α. Dual‐luciferase reporter assays were performed to assess the binding of miR‐518a‐5p to LINC01126 and HIF‐1α. RNA immunoprecipitation assay was used to identify whether LINC01126 and miR‐518a‐5p were significantly enriched in AGO‐containing micro‐ribonucleoprotein complexes.
Results
We selected LINC01126, which was the most highly expressed lncRNA, to further verify its functions in periodontitis‐induced hypoxia. The expression of LINC01126 was increased in periodontal tissues. In vitro experiment demonstrated that LINC01126 suppressed proliferation, promoted apoptosis and inflammation of hPDLCs under hypoxia via sponging miR‐518a‐5p. Moreover, we identified HIF‐1α acted as a direct target of miR‐518a‐5p in hPDLCs and LINC01126 promoted periodontitis pathogenesis by regulating the miR‐518a‐5p/HIF‐1α/MAPK pathway.
Abstract
LINC01126, which was the most highly expressed lncRNA, to further verify its functions in periodontitis‐induced hypoxia. The expression of LINC01126 were increased in periodontal tissues. In vitro experiment demonstrated that LINC01126 suppressed proliferation, promoted apoptosis and inflammation of hPDLCs under hypoxia via sponging miR‐518a‐5p. Moreover, we identified HIF‐1α acted as a direct target of miR‐518a‐5p in hPDCLs and LINC01126 promoted periodontitis pathogenesis by regulating the miR‐518a‐5p/HIF‐1α/MAPK pathway. In brief, LINC01126 promotes periodontitis pathogenesis of hPDLCs via miR‐518a‐5p/HIF‐1α/MAPK pathway, providing a possible clue for LINC01126‐based periodontal therapeutic approaches.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language?
- Record: found
- Abstract: found
- Article: not found
Non-coding RNA networks in cancer
- Record: found
- Abstract: found
- Article: not found