- Record: found
- Abstract: found
- Article: found
Multiple freeze-thaw cycles lead to a loss of consistency in poly(A)-enriched RNA sequencing

Read this article at
Abstract
Background
Both RNA-Seq and sample freeze-thaw are ubiquitous. However, knowledge about the impact of freeze-thaw on downstream analyses is limited. The lack of common quality metrics that are sufficiently sensitive to freeze-thaw and RNA degradation, e.g. the RNA Integrity Score, makes such assessments challenging.
Results
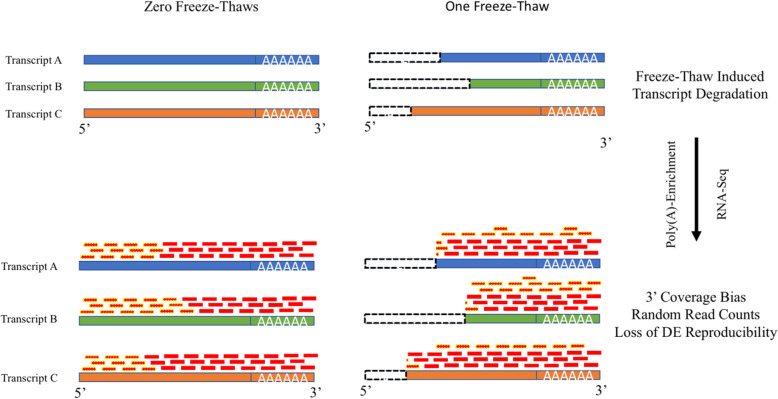
Here we quantify the impact of repeated freeze-thaw cycles on the reliability of RNA-Seq by examining poly(A)-enriched and ribosomal RNA depleted RNA-seq from frozen leukocytes drawn from a toddler Autism cohort. To do so, we estimate the relative noise, or percentage of random counts, separating technical replicates. Using this approach we measured noise associated with RIN and freeze-thaw cycles. As expected, RIN does not fully capture sample degradation due to freeze-thaw. We further examined differential expression results and found that three freeze-thaws should extinguish the differential expression reproducibility of similar experiments. Freeze-thaw also resulted in a 3′ shift in the read coverage distribution along the gene body of poly(A)-enriched samples compared to ribosomal RNA depleted samples, suggesting that library preparation may exacerbate freeze-thaw-induced sample degradation.
Related collections
Most cited references55

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found