- Record: found
- Abstract: found
- Article: found
The nuclear transportation routes of membrane-bound transcription factors
Read this article at
Abstract
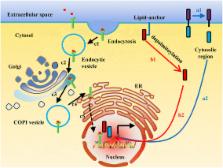
Membrane-bound transcription factors (MTFs) are transcription factors (TFs) that are anchored in membranes in a dormant state. Activated by external or internal stimuli, MTFs are released from parent membranes and are transported to the nucleus. Existing research indicates that some plasma membrane (PM)-bound proteins and some endoplasmic reticulum (ER) membrane-bound proteins have the ability to enter the nucleus. Upon specific signal recognition cues, some PM-bound TFs undergo proteolytic cleavage to liberate the intracellular fragments that enter the nucleus to control gene transcription. However, lipid-anchored PM-bound proteins enter the nucleus in their full length for depalmitoylation. In addition, some PM-bound TFs exist as full-length proteins in cell nucleus via trafficking to the Golgi and the ER, where membrane-releasing mechanisms rely on endocytosis. In contrast, the ER membrane-bound TFs relocate to the nucleus directly or by trafficking to the Golgi. In both of these pathways, only the fragments of the ER membrane-bound TFs transit to the nucleus. Several different nuclear trafficking modes of MTFs are summarized in this review, providing an effective supplement to the mechanisms of signal transduction and gene regulation. Moreover, targeting intracellular movement pathways of disease-associated MTFs may significantly improve the survival of patients.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis.
- Record: found
- Abstract: found
- Article: not found
The hardwiring of development: organization and function of genomic regulatory systems.
- Record: found
- Abstract: found
- Article: not found