- Record: found
- Abstract: found
- Article: not found
Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma
Read this article at
Abstract
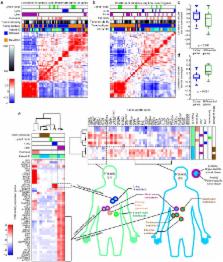
Pancreatic ductal adenocarcinoma (PDAC) remains a lethal disease with a 5-year survival of 4%. A key hallmark of PDAC is extensive stromal involvement, which makes capturing precise tumor-specific molecular information difficult. Here, we have overcome this problem by applying blind source separation to a diverse collection of PDAC gene expression microarray data, which includes primary, metastatic, and normal samples. By digitally separating tumor, stroma, and normal gene expression, we have identified and validated two tumor-specific subtypes including a “basal-like” subtype which has worse outcome, and is molecularly similar to basal tumors in bladder and breast cancer. Furthermore, we define “normal” and “activated” stromal subtypes which are independently prognostic. Our results provide new insight into the molecular composition of PDAC which may be used to tailor therapies or provide decision support in a clinical setting where the choice and timing of therapies is critical.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Oncogenic pathway signatures in human cancers as a guide to targeted therapies.
- Record: found
- Abstract: found
- Article: not found
Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin.
- Record: found
- Abstract: found
- Article: not found
