- Record: found
- Abstract: found
- Article: not found
MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data
Read this article at
Abstract
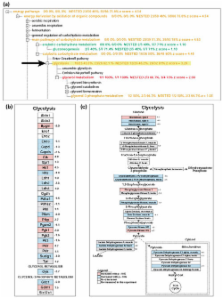
MAPPFinder is a tool that creates a global gene-expression profile across all areas of biology by integrating the annotations of the Gene Ontology (GO) Project with the free software package GenMAPP.
Abstract
MAPPFinder is a tool that creates a global gene-expression profile across all areas of biology by integrating the annotations of the Gene Ontology (GO) Project with the free software package GenMAPP http://www.GenMAPP.org. The results are displayed in a searchable browser, allowing the user to rapidly identify GO terms with over-represented numbers of gene-expression changes. Clicking on GO terms generates GenMAPP graphical files where gene relationships can be explored, annotated, and files can be freely exchanged.
Related collections
Most cited references16
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
Creating the gene ontology resource: design and implementation.
- Record: found
- Abstract: not found
- Article: not found
